Introduction
Alignment of large-scale serial-section electron microscopy (ssEM) images is crucial for successful analysis in nano-scale connectomics. Despite various image registration algorithms proposed in the past, large-scale ssEM alignment remains challenging due to the size and complex nature of the data.
Recently, the application of unsupervised machine learning in medical image registration has shown promise in efforts to replace an expensive numerical computation process with a once-deployed feed-forward neural network. However, the anisotropy in most ssEM data makes it difficult to directly adopt such learning-based methods for the registration of these images.
Weakly Supervised Learning in Deformable EM Image Registration using Slice Interpolation
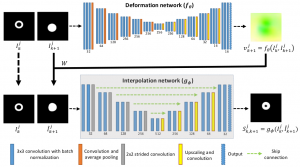
we propose a novel deformable image registration approach based on weakly supervised learning that can be applied to registering ssEM images at scale. The proposed method leverages slice interpolation to improve the registration between images with sudden and large structural changes.
Results
Mouse cerebellum dataset alignment results. Upper row: baseline data prealigned using AlignTK; lower row: improved alignment using our method.
 [bibtex file=thanh_isbi_2019.bib format=plain
[bibtex file=thanh_isbi_2019.bib format=plain
process_titles=0]



